Note
This is a community plugin, an external project maintained by its respective author. Community plugins are not part of FiftyOne core and may change independently. Please review each plugin’s documentation and license before use.
Implementing MedGemma 1.5 as a Remote Zoo Model for FiftyOne#
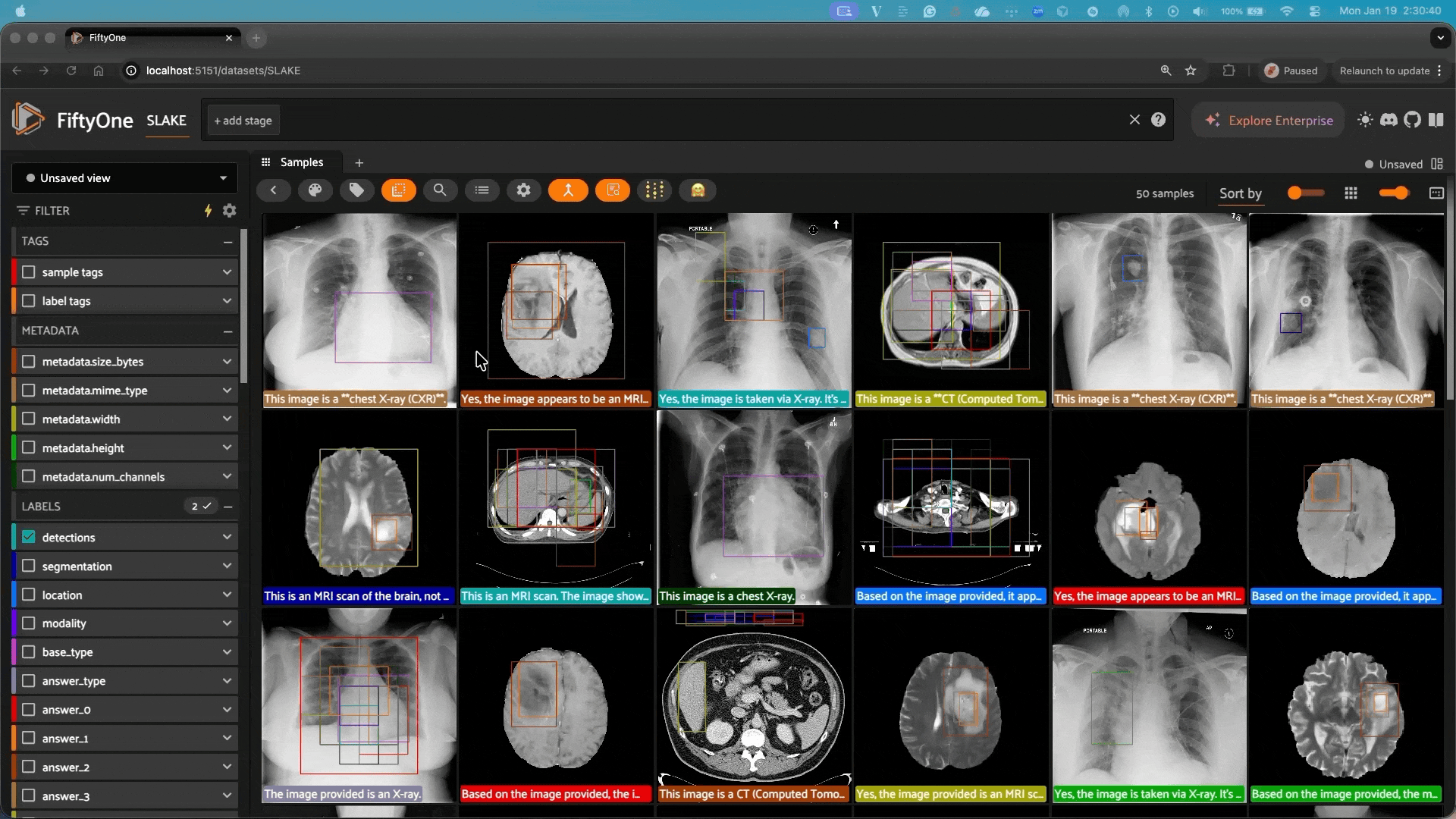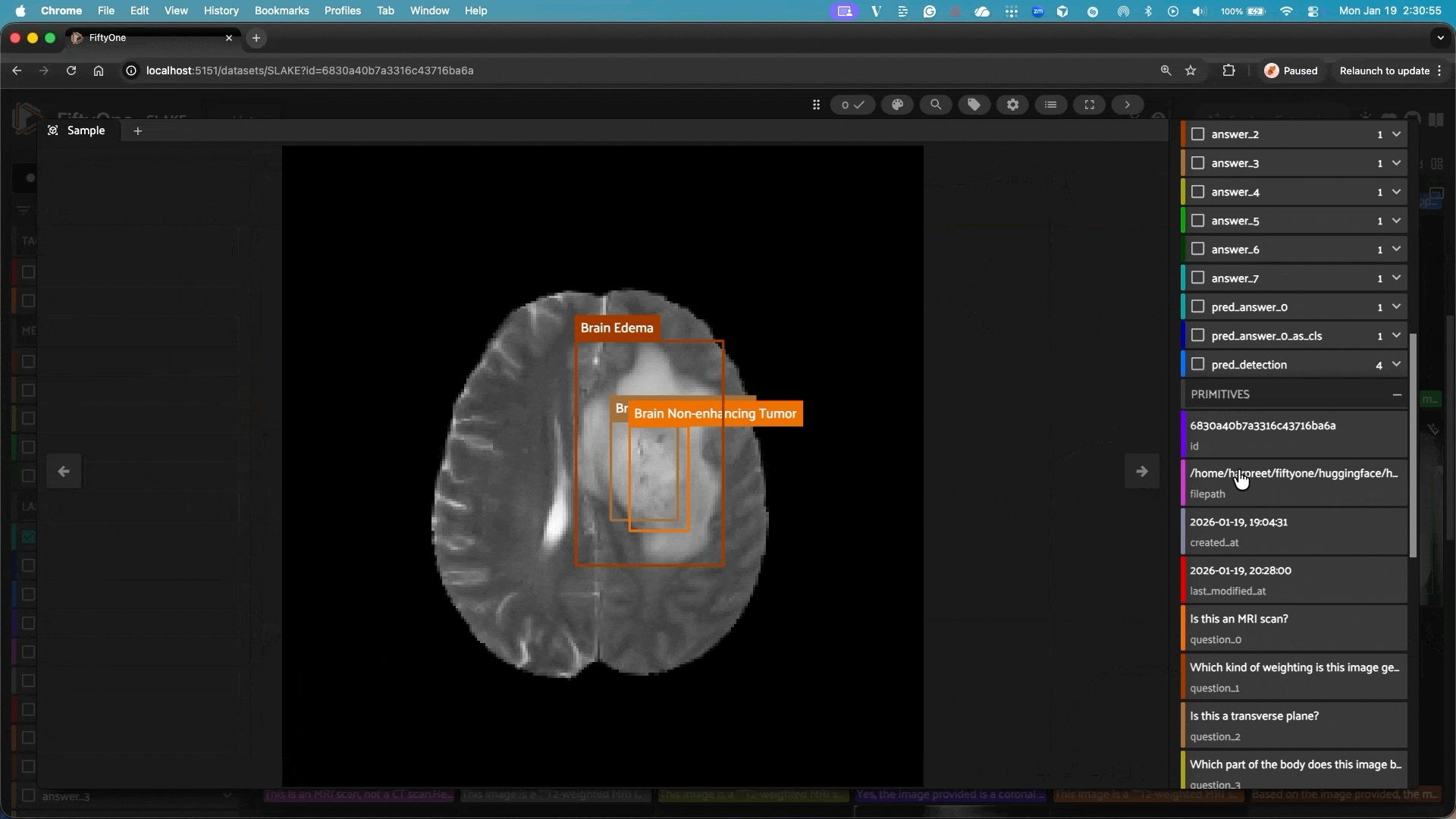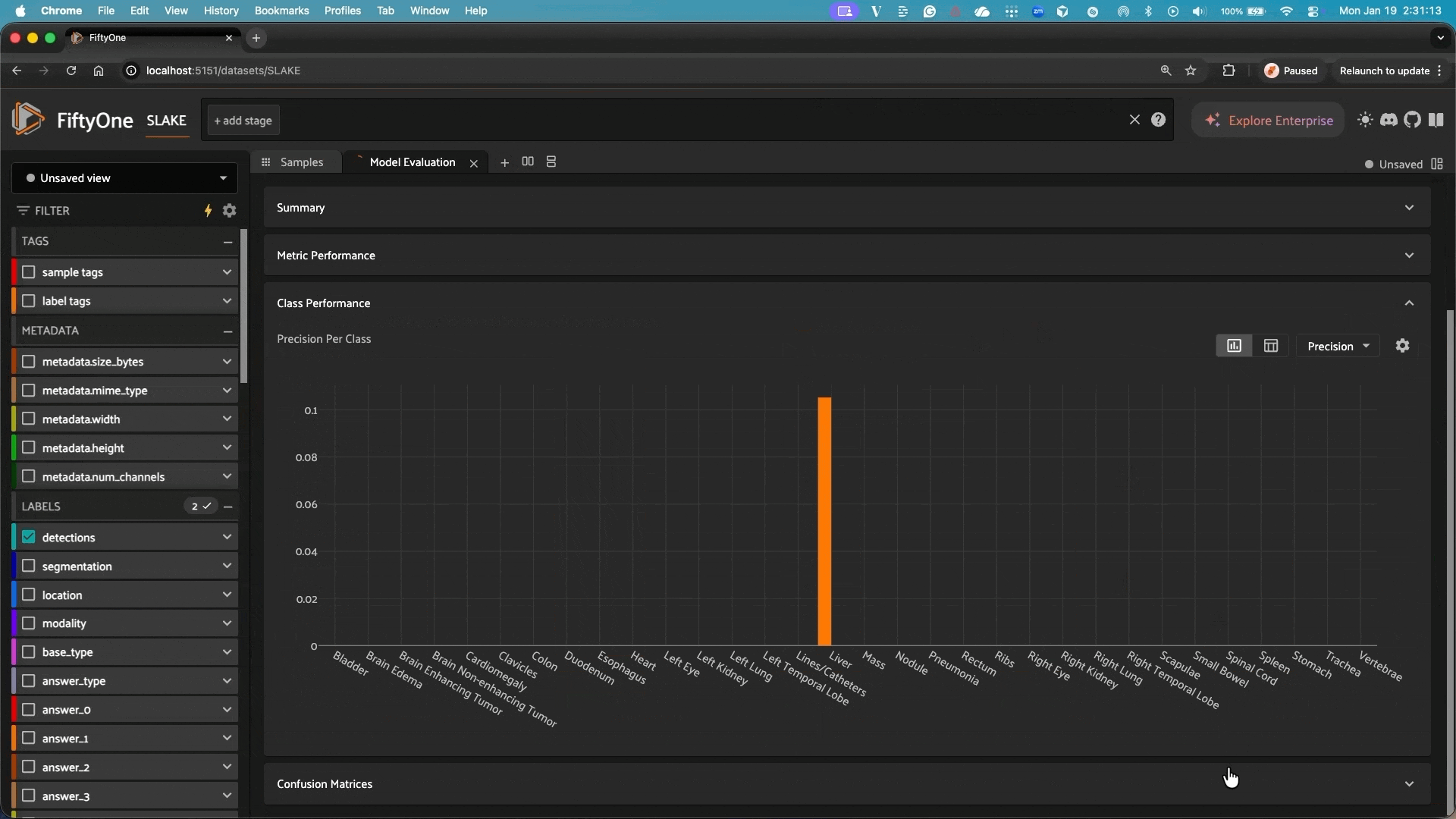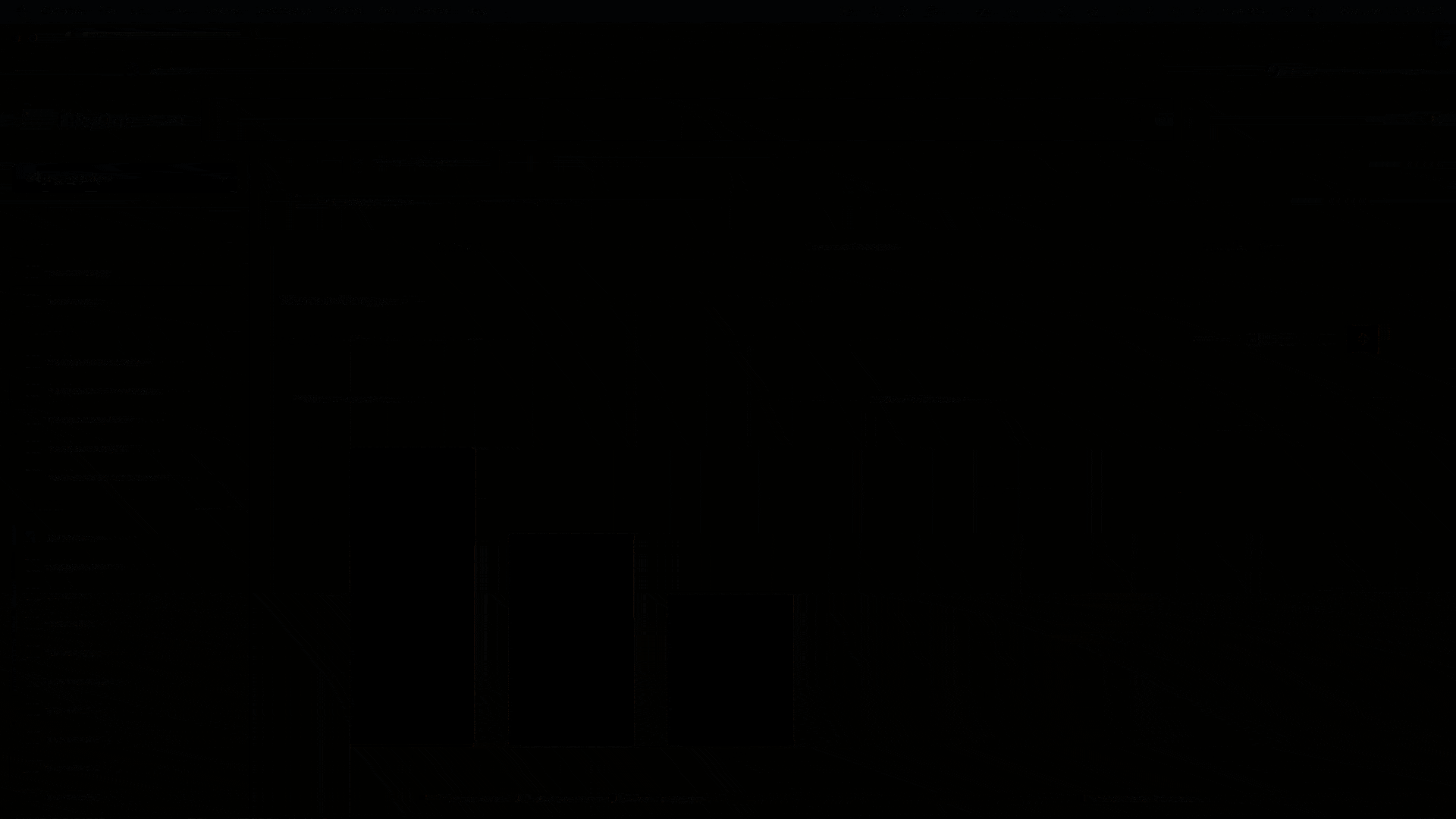
This repository integrates Google’s MedGemma models with FiftyOne, allowing you to easily use these powerful medical AI models for analyzing and classifying medical images in your FiftyOne datasets.
ℹ Important! Be sure to request access to the model!#
This is a gated model, so you will need to fill out the form on the model card: https://huggingface.co/google/medgemma-1.5-4b-it
Approval should be instantaneous.
You’ll also have to set your Hugging Face in your enviornment:
export HF_TOKEN="your_token"
Or sign-in to Hugging Face via the CLI:
hf auth login
What is MedGemma?#
MedGemma is a collection of Gemma 3 variants that are trained specifically for medical text and image comprehension. These models excel at understanding various medical imaging modalities including:
Chest X-rays
Dermatology images
Ophthalmology images
Histopathology slides
This integration is for MedGemma 1.5 4B, a multimodal version that can process both images and text
Installation#
First, ensure you have FiftyOne installed:
pip install fiftyone
Then register this repository as a custom model source:
import fiftyone.zoo as foz
foz.register_zoo_model_source("https://github.com/harpreetsahota204/medgemma_1_5", overwrite=True)
Usage#
Download and Load the Model#
import fiftyone.zoo as foz
# Download the model (only needed once)
foz.download_zoo_model(
"https://github.com/harpreetsahota204/medgemma_1_5",
model_name="google/medgemma-1.5-4b-it",
)
# Load the model
model = foz.load_zoo_model(
"google/medgemma-1.5-4b-it"
)
Setting the Operation Mode#
The model supports three main operations:
# For medical image classification
model.operation = "classify"
# For visual question answering on medical images
model.operation = "vqa"
# For anatomical structure and pathology detection
model.operation = "detect"
Classification Example#
import fiftyone as fo
from fiftyone.utils.huggingface import load_from_hub
dataset = load_from_hub(
"Voxel51/MedXpertQA",
name="MedXpertQA",
max_samples=10,
overwrite=True
)
# Set classification parameters
model.operation = "classify"
model.prompt = "What medical conditions are visible in this image?"
# Run classification on the dataset
dataset.apply_model(model, label_field="medgemma_classifications")
Visual Question Answering Example#
# Set VQA parameters
model.operation = "vqa"
model.prompt = "Is there evidence of pneumonia in this chest X-ray? Explain your reasoning."
# Apply to dataset
dataset.apply_model(model, label_field="pneumonia_assessment")
Detection Example#
Localize anatomical structures and pathologies in medical images. The model outputs bounding boxes in FiftyOne’s Detections format.
# Set detection mode
model.operation = "detect"
# Get labels to detect (e.g., from ground truth)
labels = dataset.distinct("detections.detections.label")
labels_str = ", ".join(labels)
# Prompt for localization
model.prompt = f"""Locate the following in this scan: {labels_str}.
Output the final answer in the format "Final Answer: X" where X is a JSON list of objects.
The object needs a "box_2d" and "label" key.
If the object is not present in the scan, skip it and don't output anything for that object.
Answer:"""
# Apply detection
dataset.apply_model(
model,
label_field="pred_detection",
batch_size=8,
num_workers=4,
)
Note: Detection mode automatically applies square padding to preserve aspect ratio, ensuring bounding box coordinates map correctly between model output and the original image.
Batching for Faster Inference#
MedGemma supports efficient batch processing for significantly faster inference on large datasets. Use batch_size and num_workers parameters:
Global Prompt with Batching#
Set a single prompt that applies to all samples:
# Get unique labels from your dataset
body_system_labels = dataset.distinct("modality.label")
model.operation = "classify"
model.prompt = "As a medical expert your task is to classify this image into exactly one of the following types: " + ", ".join(body_system_labels)
# Apply with batching for faster inference
dataset.apply_model(
model,
label_field="pred_modality",
batch_size=64,
num_workers=8,
)
Per-Sample Prompts with Batching#
Use prompt_field to read prompts from a field on each sample:
model.operation = "vqa"
# Each sample uses its own prompt from the "question" field
dataset.apply_model(
model,
label_field="pred_answer",
prompt_field="question",
batch_size=64,
num_workers=8,
)
This is useful when you have different questions or prompts stored per sample in your dataset.
Custom System Prompts#
You can customize the system prompt to better suit your specific needs:
model.system_prompt = """You are an expert dermatologist specializing in skin cancer detection.
Analyze the provided skin lesion image and determine if there are signs of malignancy.
Provide your assessment in JSON format with detailed observations."""
Performance Considerations#
For optimal performance, a CUDA-capable GPU is recommended
Use
batch_sizeandnum_workersparameters inapply_model()for significantly faster inference on large datasetsTypical settings:
batch_size=32-64,num_workers=4-8(adjust based on your GPU memory)
Example notebook#
You can refer to the example notebook to get hands on.
License#
MedGemma is governed by the Health AI Developer Foundations terms of use.
This integration is licensed under the Apache 2.0 License.
Notes#
This integration is designed for research and development purposes
Always validate model outputs in clinical contexts
Review the MedGemma documentation for detailed information about the model’s capabilities and limitations
Citation#
If you use MedGemma in your research, please cite the MedGemma Technical Report:
@article{sellergren2025medgemma,
title={MedGemma Technical Report},
author={Sellergren, Andrew and Kazemzadeh, Sahar and Jaroensri, Tiam and Kiraly, Atilla and Traverse, Madeleine and Kohlberger, Timo and Xu, Shawn and Jamil, Fayaz and Hughes, Cían and Lau, Charles and others},
journal={arXiv preprint arXiv:2507.05201},
year={2025}
}
