 Run in Google Colab Run in Google Colab
|
 View source on GitHub View source on GitHub
|
|
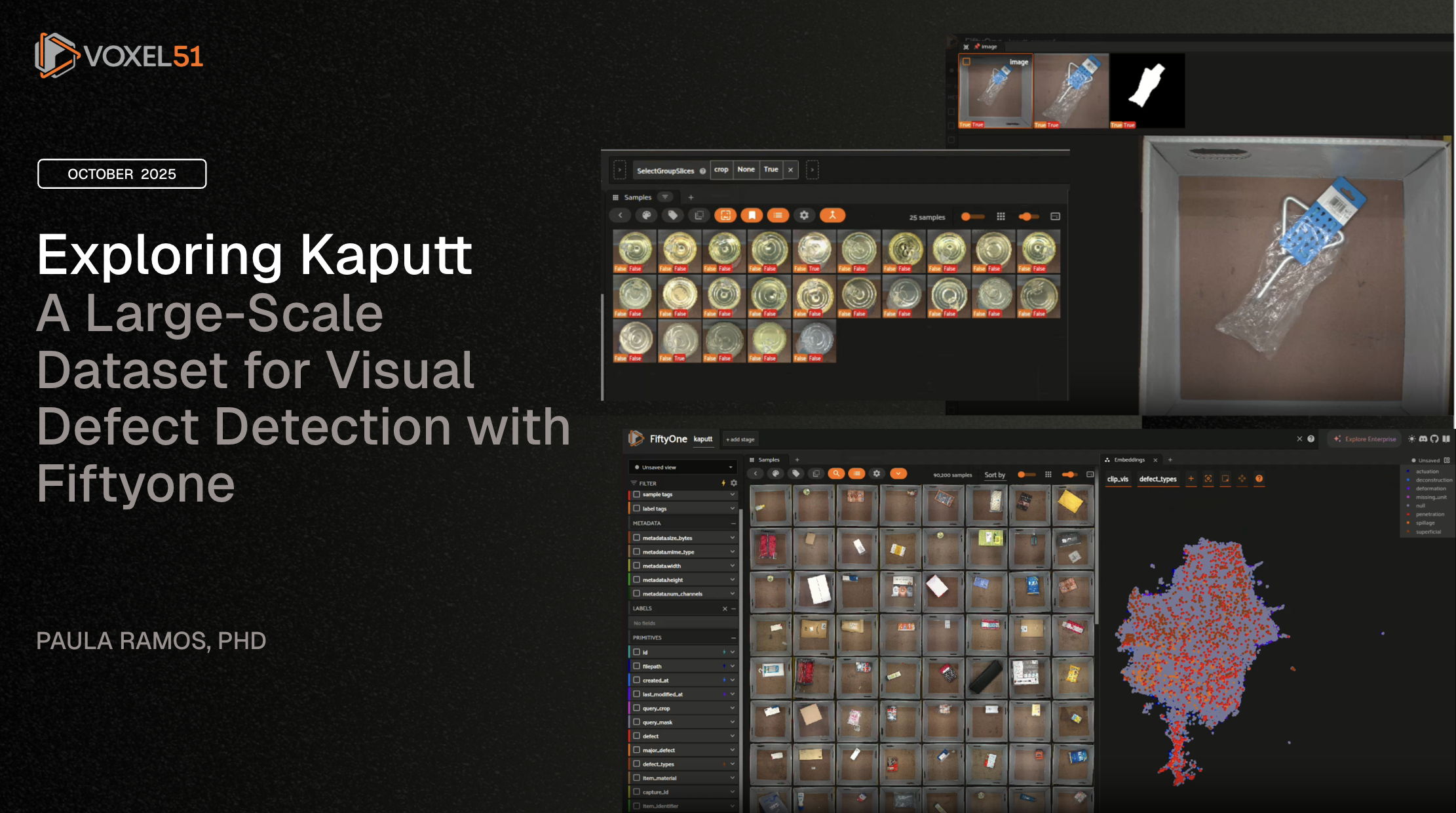
Exploring Kaputt Dataset#
The Kaputt dataset marks a new era for visual defect detection in Retail Logistics. Developed by researchers from Amazon and the University of Oxford, Kaputt contains more than 238,000 images and 48,000 unique products, including over 29,000 defective instances. It is 40x larger than existing benchmarks such as MVTec-AD and VisA.
While previous datasets focused on tightly controlled manufacturing settings, Kaputt introduces real-world complexity, products with varying shapes, materials, lighting conditions, and poses. This makes defect detection far more challenging and realistic: even state-of-the-art models struggle to exceed 56.9% AUROC on this benchmark.
So, what’s the takeaway?
Load and explore the Kaputt dataset interactively using FiftyOne.
Visualize its structure and metadata fields.
Compute and analyze embeddings, find similar samples query and filter the dataset from different inputs.
Experiment with VLMs as FastVLM and other models from the FiftyOne Model Zoo.
Demonstrate how FiftyOne helps uncover valuable insights into data quality, bias, and model performance.
Setup#
If you haven’t already, install the required packages. These cells will only install packages if they’re missing. This notebook was tested in a Python Env (py 3.10)
[ ]:
!pip install fiftyone umap-learn torch torchvision pandas pyarrow
Request access and download the dataset locally and dataset structure#
Fill the Request Form
Locate the access form for the Kaputt dataset, available on the dataset’s official website or publication page.
Complete all required fields with accurate information such as your name, email, and affiliation.
Submit the Form
Double-check your details before submission.
Click Submit to send your access request.
Check Your Email
Open the inbox of the email address you provided.
Look for a confirmation or dataset access message.
Dataset structure#
In this notebook we will explore the complete query folder, for the other folders you can extend with the provided code.
kaputt/
├── datasets/ # Parquet metadata (index tables)
│ ├── query-train.parquet
│ ├── query-validation.parquet
│ ├── query-test.parquet
│ ├── reference-train.parquet
│ ├── reference-validation.parquet
│ ├── reference-test.parquet
│ └── README.md
│
├── query-image/ # Full query images (main inputs)
│ └── data/
│ ├── train/
│ │ └── query-data/image/
│ │ ├── <capture_id>.jpg
│ │ ├── ...
│ ├── validation/
│ │ └── query-data/image/
│ │ ├── <capture_id>.jpg
│ │ ├── ...
│ └── test/
│ └── query-data/image/
│ ├── <capture_id>.jpg
│ ├── ...
│
├── query-crop/ # Cropped item regions
│ └── data/
│ ├── train/
│ │ └── query-data/crop/
│ │ ├── <capture_id>.jpg
│ │ ├── ...
│ ├── validation/
│ │ └── query-data/crop/
│ │ ├── <capture_id>.jpg
│ │ ├── ...
│ └── test/
│ └── query-data/crop/
│ ├── <capture_id>.jpg
│ ├── ...
│
├── query-mask/ # Binary/segmentation masks
│ └── data/
│ ├── train/
│ │ └── query-data/mask/
│ │ ├── <capture_id>.png
│ │ ├── ...
│ ├── validation/
│ │ └── query-data/mask/
│ │ ├── <capture_id>.png
│ │ ├── ...
│ └── test/
│ └── query-data/mask/
│ ├── <capture_id>.png
│ ├── ...
│
├── reference-image/ # Reference (non-defective) images
│ └── data/
│ ├── train/reference-data/image/
│ ├── validation/reference-data/image/
│ └── test/reference-data/image/
│
├── reference-crop/ # Crops for reference images
│ └── data/
│ ├── train/reference-data/crop/
│ ├── validation/reference-data/crop/
│ └── test/reference-data/crop/
│
├── reference-mask/ # Segmentation masks for reference images
│ └── data/
│ ├── train/reference-data/mask/
│ ├── validation/reference-data/mask/
│ └── test/reference-data/mask/
│
├── sample-data/ # Small subset for testing
│ ├── data/
│ │ └── train/
│ │ ├── query-data/
│ │ │ ├── image/
│ │ │ ├── crop/
│ │ │ └── mask/
│ │ └── reference-data/
│ │ ├── image/
│ │ ├── crop/
│ │ └── mask/
│ ├── query-sample.parquet
│ └── reference-sample.parquet
│
└── kaputt-release/ # Original release version (mirrors structure above)
├── train/
│ ├── query-data/
│ └── reference-data/
├── validation/
│ ├── query-data/
│ └── reference-data/
└── test/
├── query-data/
└── reference-data/
Import Kaputt (Query Only)#
This cell imports the query portion of the Kaputt dataset into FiftyOne. It reads the query Parquet files from /datasets/, builds absolute paths for images, crops, and masks, and creates a FiftyOne dataset with fields for defect attributes and item metadata. Only train and validation splits are loaded for faster testing.
[ ]:
# import_kaputt_to_fiftyone_queries_only.py
#
# Media layout:
# <ROOT>/query-image/data/<split>/query-data/image/<filename>
# <ROOT>/query-crop/data/<split>/query-data/crop/<filename>
# <ROOT>/query-mask/data/<split>/query-data/mask/<filename>
#
# Parquets live in <ROOT>/datasets/ with names like:
# query-train.parquet, query_validation.parquet, query-train.parquet.gz, etc.
#
# Loads ONLY query media (+ attrs) for splits: train, validation
import os
from pathlib import Path
import glob
import pandas as pd
import fiftyone as fo
# ---------- CONFIG ----------
EXTERNAL_ROOT = "/your/main/folder/for/kaputt" # <-- your kaputt root
DATASET_NAME = "kaputt"
PARQUET_DIR = "datasets"
SPLITS = ("train", "validation") # <-- test excluded
NUM_WORKERS_METADATA = 8
# ---------------------------
root = Path(EXTERNAL_ROOT).resolve()
pdir = (root / PARQUET_DIR).resolve()
# kind -> (top media folder, terminal folder under query-data)
# Final path: <root>/<media_folder>/data/<split>/query-data/<terminal>/<filename>
MEDIA_MAPPING = {
"query_image": ("query-image", "image"),
"query_crop": ("query-crop", "crop"),
"query_mask": ("query-mask", "mask"),
}
def _basename(pathlike) -> str | None:
if pathlike is None or (isinstance(pathlike, float) and pd.isna(pathlike)):
return None
s = str(pathlike).replace("\\", "/").rstrip("/")
base = os.path.basename(s)
return base or None
def resolve_path_from_schema(rel, split: str, kind: str) -> str | None:
"""
Build absolute path:
<root>/<media_folder>/data/<split>/query-data/<terminal>/<filename>
rel may be absolute, relative, or just a filename.
"""
if rel is None or (isinstance(rel, float) and pd.isna(rel)):
return None
if os.path.isabs(str(rel)) and os.path.exists(str(rel)):
return str(Path(rel).resolve())
fname = _basename(rel)
if not fname:
return None
media_folder, terminal = MEDIA_MAPPING[kind]
abs_path = root / media_folder / "data" / split / "query-data" / terminal / fname
return str(abs_path)
def find_query_parquet(split: str) -> Path:
"""
Robustly find a parquet file for a split inside <root>/datasets/.
Supports common variants and .parquet.gz.
"""
candidates = []
patterns = [
f"query-{split}.parquet",
f"query_{split}.parquet",
f"query-*{split}*.parquet",
f"query-{split}.parquet.gz",
f"query_{split}.parquet.gz",
f"query-*{split}*.parquet.gz",
]
for pat in patterns:
candidates.extend(glob.glob(str(pdir / pat)))
candidates = sorted(set(candidates))
if not candidates:
raise FileNotFoundError(
f"No query parquet found for split '{split}'. "
f"Tried under: {pdir}\nPatterns: {patterns}"
)
preferred = pdir / f"query-{split}.parquet"
if preferred.exists():
return preferred
return Path(candidates[0])
def read_parquet_robust(fp: Path) -> pd.DataFrame:
try:
return pd.read_parquet(fp)
except Exception as e:
raise RuntimeError(
f"Failed to read parquet file: {fp}\n"
f"Error: {type(e).__name__}: {e}\n"
f"Tip: ensure it's a valid parquet (or parquet.gz) file."
) from e
def prepare_split(split: str):
try:
q_fp = find_query_parquet(split)
except FileNotFoundError as e:
print(f"[INFO] Skipping split '{split}': {e}")
return []
print(f"[INFO] Using query parquet for '{split}': {q_fp}")
q = read_parquet_robust(q_fp)
# Normalize optional columns
for col in ("defect", "major_defect", "defect_types"):
if col not in q.columns:
q[col] = None
samples, skipped_missing = [], 0
for _, row in q.iterrows():
query_image = resolve_path_from_schema(row.get("query_image"), split, "query_image")
query_crop = resolve_path_from_schema(row.get("query_crop"), split, "query_crop")
query_mask = resolve_path_from_schema(row.get("query_mask"), split, "query_mask")
# Require main media
if not (query_image and os.path.exists(query_image)):
skipped_missing += 1
continue
# defect_types -> list
dtypes = row.get("defect_types", None)
if isinstance(dtypes, str):
dtypes = [s.strip() for s in dtypes.split(",") if s.strip()]
elif dtypes is None or (isinstance(dtypes, float) and pd.isna(dtypes)):
dtypes = []
s = fo.Sample(filepath=query_image, tags=[split])
if query_crop and os.path.exists(query_crop):
s["query_crop"] = query_crop
if query_mask and os.path.exists(query_mask):
s["query_mask"] = query_mask
s["defect"] = bool(row.get("defect", False))
s["major_defect"] = bool(row.get("major_defect", False))
s["defect_types"] = dtypes
s["item_material"] = row.get("item_material", None)
s["capture_id"] = row.get("capture_id", None)
s["item_identifier"] = row.get("item_identifier", None)
s["split"] = split
samples.append(s)
if skipped_missing:
print(f"[WARN] Split '{split}': skipped {skipped_missing} samples with missing query_image")
return samples
Create and Save FiftyOne Dataset#
This cell creates a new FiftyOne dataset named after DATASET_NAME, adds samples for each available split (train and validation), and computes image metadata (dimensions, channels, etc.). If a dataset with the same name already exists, it is replaced to ensure a clean import.
After adding all samples:
The dataset is saved and marked as persistent.
Split counts are printed for verification.
Metadata is computed in parallel using
NUM_WORKERS_METADATA
[ ]:
if fo.dataset_exists(DATASET_NAME):
fo.delete_dataset(DATASET_NAME)
ds = fo.Dataset(DATASET_NAME)
total_added = 0
for split in SPLITS:
samples = prepare_split(split)
if samples:
ds.add_samples(samples)
print(f"[INFO] Added {len(samples)} samples for split '{split}'")
total_added += len(samples)
ds.save()
print(ds)
try:
print("Counts by split:", ds.count_values("split"))
except Exception as e:
print("[INFO] Could not aggregate counts by split:", e)
if len(ds) > 0:
try:
print("[INFO] Computing metadata...")
ds.compute_metadata(overwrite=True, num_workers=NUM_WORKERS_METADATA)
print("[INFO] Done computing metadata.")
except Exception as e:
print(f"[WARN] Failed to compute metadata: {e}")
else:
print("[WARN] Dataset is empty; skipped metadata computation")
ds.persistent = True
print(f"[INFO] Dataset '{DATASET_NAME}' is ready with {total_added} samples.")
Compute CLIP Embeddings and Similarity Index#
This cell uses the CLIP ViT-B/32 model from the FiftyOne Model Zoo to generate visual embeddings for all samples in the dataset. The embeddings are stored in the field clip_embeddings and used to build a similarity index (clip_sim) via the FiftyOne Brain module.
This enables efficient image similarity search, semantic clustering, and content-based exploration directly within FiftyOne.
[ ]:
import fiftyone.zoo as foz
import fiftyone.brain as fob
# Load CLIP model
model = foz.load_zoo_model("clip-vit-base32-torch")
# Compute embeddings for all samples
ds.compute_embeddings(model, embeddings_field="clip_embeddings")
# Create similarity index from pre-computed embeddings
fob.compute_similarity(
ds,
model="clip-vit-base32-torch",
embeddings="clip_embeddings",
brain_key="clip_sim",
)
Dimensionality Reduction with UMAP#
This cell applies UMAP (Uniform Manifold Approximation and Projection) to the CLIP embeddings stored in clip_embeddings. It computes a 2D visualization of the dataset’s feature space using the FiftyOne Brain module and saves it under the key clip_vis.
This allows interactive exploration of the dataset in embedding space, revealing visual clusters and relationships between samples.
[ ]:
# Dimensionality reduction using UMAP on the embeddings
fob.compute_visualization(
ds,
embeddings="clip_embeddings",
method="umap",
brain_key="clip_vis",
)
Create Index for Faster Filtering#
This cell creates a compound index on the fields defect_types, item_material, and split to optimize query performance in FiftyOne. By indexing these commonly filtered fields, dataset operations such as searching, filtering, and aggregating by defect category or material type become significantly faster, especially when working with large datasets.
[ ]:
# Create an index to speed up filtering by defect types, item material, and split
ds.create_index(
[("defect_types", 1), ("item_material", 1), ("split", 1)]
)
Open a web browser session to play interactively with your dataset, metadata and embeddings.
Explore visually in the FiftyOne App#
[ ]:
# Launch FiftyOne App
session = fo.launch_app(ds, port=5151, auto=False)
session.wait()
Apply FastVLM to Evaluate Defect Severity#
This cells integrate the FastVLM model from the open-source community into FiftyOne to analyze the Kaputt dataset. It registers the model source from GitHub, downloads the desired FastVLM variant (0.5B, 1.5B, or 7B), and loads it into the environment.
Using a detailed prompt adapted from the model authors, the system asks the model to reason about item condition and defect severity. The model’s output is stored in a new field called result, containing structured JSON with:
"condition" → "DAMAGED" or "UNDAMAGED"
"severity" → Numeric score from 0 (pristine) to 10 (completely destroyed)
This enables analysis of how a large vision-language model interprets real-world packaging damage across the Kaputt dataset.
[ ]:
import fiftyone.zoo as foz
# Register the model source
foz.register_zoo_model_source(
"https://github.com/harpreetsahota204/fast_vlm",
overwrite=True
)
# Download the desired model variant (first time only)
# Choose from: "apple/FastVLM-0.5B", "apple/FastVLM-1.5B", or "apple/FastVLM-7B"
foz.download_zoo_model(
"https://github.com/harpreetsahota204/fast_vlm",
model_name="apple/FastVLM-7B" # Change to desired model variant
)
Add predictions to your dataset#
[ ]:
# Load the model
model = foz.load_zoo_model("apple/FastVLM-1.5B")
# Generate creative content with a custom prompt
model.prompt = ("You are a highly skilled subject matter expert "
"for inventory quality assurance and control. The presented "
"image shows an item inside a tray. You have to determine whether "
"the item is in pristine condition and can be sold as new and shipped "
"to the customer as is, or whether it is damaged in any way and needs "
"further attention before it can be shipped. Consider the "
"following damage categories: crushed, tear, hole, deformed, ripped, "
"deconstructed. Typical defects also include open boxes, or damaged and ripped packaging. "
"Sometimes if the packaging is damaged, the item itself may become deconstructed and parts "
"of the content may fall out. The container itself may be dirty "
"which should not count as damage. However, if there is "
"spillage that originated from a liquid item, then it must "
"be called out as damage. Pay close attention to books "
"and especially to corners of front or back pages. Moreover, "
"items that a deconstructed, i.e. where the original "
"packaging is damaged or fell off, should be flagged as "
"damaged. In addition to the final decision specified by "
"DAMAGED or UNDAMAGED, please also provide "
"the severity score on a scale from 0 (pristine condition) "
"to 10 (completely destroyed). Think step-by-step and "
"provide the final response as json with keys 'condition' "
"and 'severity'."
)
ds.apply_model(model, label_field="result")
[ ]:
session = fo.launch_app(ds, port=5151, auto=False)
Creating a Grouped Dataset with Images, Crops, and Masks in FiftyOne#
This code demonstrates how to create a grouped dataset in FiftyOne, where each group can contain up to three related slices: the original image, a crop, and a mask. This structure is useful for organizing and visualizing multimodal or multiview data, such as associating each image with its corresponding crop and segmentation mask.
Key steps:
Dataset Setup:
Checks if a grouped dataset with the specified name exists and deletes it if so.
Creates a new grouped dataset and adds a group field (with
"image"as the default slice).
Sample Processing:
Iterates through each sample in the original dataset.
Copies relevant fields (excluding system fields) to new grouped samples.
Image Slice:
Always adds the original image as a slice in the group.
Crop Slice:
If a crop is available (
sample.query_crop), adds it as a separate slice.
Mask Slice:
If a mask is available (
sample.query_mask), reads and normalizes it to the 0-255 range.Applies a binary threshold to create a binarized mask.
Only adds the mask slice if it contains meaningful (non-black) data.
Debugging and Statistics:
Tracks and prints the number of total samples processed, masks added, and masks skipped.
Prints a summary of the grouped dataset, including the number of mask samples.
This approach leverages FiftyOne’s native grouping feature, which is ideal for paired or multimodal data exploration and visualization. For more details on grouped datasets and their use cases, see the FiftyOne grouped datasets documentation and related example notebooks.
Grouped datasets in FiftyOne allow you to organize related samples (such as images, masks, and crops) under a common group, enabling synchronized visualization and analysis across different data modalities or views.
[ ]:
import cv2
import numpy as np
# Delete and recreate the grouped dataset
grouped_dataset_name = "kaputt_grouped"
if grouped_dataset_name in fo.list_datasets():
fo.delete_dataset(grouped_dataset_name)
grouped_dataset = fo.Dataset(grouped_dataset_name, persistent=True, overwrite=True)
grouped_dataset.add_group_field("group", default="image")
grouped_samples = []
# Counters for debugging
total_samples = 0
masks_added = 0
masks_skipped = 0
def normalize_mask(mask_img):
"""Normalize mask values to 0-255 range"""
if mask_img is None or mask_img.size == 0:
return None
min_val = mask_img.min()
max_val = mask_img.max()
# If mask is all zeros or constant, return None
if min_val == max_val:
return None
# Normalize to 0-255
normalized = ((mask_img - min_val) / (max_val - min_val) * 255).astype(np.uint8)
return normalized
for sample in ds.iter_samples(progress=True):
total_samples += 1
group = fo.Group()
# Prepare fields to copy (excluding system fields)
fields_to_copy = [
f for f in sample.field_names
if f not in ("id", "filepath", "group", "tags", "metadata")
]
# --- Original image slice ---
image_sample = fo.Sample(
filepath=sample.filepath,
group=group.element("image"),
)
for field in fields_to_copy:
image_sample[field] = sample[field]
if sample.metadata is not None:
image_sample.metadata = sample.metadata
# --- Crop slice (if available) ---
crop_sample = None
if sample.query_crop:
crop_sample = fo.Sample(
filepath=sample.query_crop,
group=group.element("crop"),
)
for field in fields_to_copy:
crop_sample[field] = sample[field]
# --- Mask slice (if available and valid) ---
mask_sample = None
if sample.query_mask:
mask_img = cv2.imread(sample.query_mask, cv2.IMREAD_GRAYSCALE)
if mask_img is not None:
# First normalize the mask to 0-255 range
normalized_mask = normalize_mask(mask_img)
if normalized_mask is not None:
# Now apply threshold on normalized mask
_, bin_mask = cv2.threshold(normalized_mask, 127, 255, cv2.THRESH_BINARY)
# Check if the mask contains any white pixels
if np.any(bin_mask):
mask_dir = os.path.join(os.path.dirname(sample.query_mask), "normalized_binarized_masks")
os.makedirs(mask_dir, exist_ok=True)
mask_filename = os.path.basename(sample.query_mask)
bin_mask_path = os.path.join(mask_dir, mask_filename)
cv2.imwrite(bin_mask_path, bin_mask)
mask_sample = fo.Sample(
filepath=bin_mask_path,
group=group.element("mask"),
)
for field in fields_to_copy:
mask_sample[field] = sample[field]
masks_added += 1
else:
masks_skipped += 1
print(f"[DEBUG] Skipped mask (all black after threshold): {sample.query_mask}")
else:
masks_skipped += 1
print(f"[DEBUG] Skipped mask (constant values): {sample.query_mask}")
else:
masks_skipped += 1
print(f"[DEBUG] Could not read mask: {sample.query_mask}")
# Add all available slices to the group
for s in (image_sample, crop_sample, mask_sample):
if s is not None:
grouped_samples.append(s)
# Add all grouped samples to the new dataset
grouped_dataset.add_samples(grouped_samples)
# Print summary
print(f"\n[INFO] Processing complete:")
print(f" Total samples processed: {total_samples}")
print(f" Masks successfully added: {masks_added}")
print(f" Masks skipped: {masks_skipped}")
print(f" Total grouped samples created: {len(grouped_samples)}")
# Verify the grouped dataset
print(f"\n[INFO] Grouped dataset info:")
print(f" Total samples: {len(grouped_dataset)}")
print(f" Group slices: {grouped_dataset.group_slices}")
# Check how many mask samples were added
mask_count = len(grouped_dataset.match({"group.name": "mask"}))
print(f" Mask samples in dataset: {mask_count}")
[ ]:
grouped_dataset.persistent = True
[ ]:
session2 = fo.launch_app(grouped_dataset, port=5152, auto=False)
Computing Embeddings and Similarity Index for Crop Slices in FiftyOne#
This code demonstrates how to compute embeddings for the “crop” slices in a grouped dataset and then create a similarity index for these crops using the CLIP model from the FiftyOne Model Zoo.
Workflow:
- Select Crop Slices:Use
select_group_slices("crop")to create a flattened view containing only the crop samples from your grouped dataset. - Load CLIP Model:Load the
"clip-vit-base32-torch"model from the FiftyOne Model Zoo, which supports generating embeddings for images and patches see: Model Zoo API Reference. - Compute Embeddings:Compute embeddings for all crop samples and store them in the
"crop_embeddings"field. This can be done using thecompute_embeddings()method, which works with any model that exposes embeddings see: Model Zoo API Reference. - Create Similarity Index:Use
compute_similarity()to create a similarity index over the crop samples, specifying the model and the field containing the precomputed embeddings. This enables similarity search and sorting by similarity for the crop slices see: Creating an index.
[ ]:
import fiftyone.zoo as foz
import fiftyone.brain as fob
# Get all crop samples as a flattened view
crop_view = grouped_dataset.select_group_slices("crop")
print(f"[INFO] Crop samples: {len(crop_view)}")
# Compute embeddings on crop samples
model = foz.load_zoo_model("clip-vit-base32-torch")
crop_view.compute_embeddings(model, embeddings_field="crop_embeddings")
# Create similarity index for crops
fob.compute_similarity(
crop_view,
model="clip-vit-base32-torch",
embeddings="crop_embeddings",
brain_key="crop_sim",
)
print("[INFO] Crop embeddings computed successfully!")
Visualizing Embeddings for Crop Slices in FiftyOne#
This code demonstrates how to compute and visualize embeddings for the “crop” slices of a grouped dataset using FiftyOne Brain and the CLIP model.
Steps:
- Select Crop Slices:Use
select_group_slices("crop")to obtain a flattened view containing only the crop samples from your grouped dataset. - Compute Embeddings and Visualization:Call
fob.compute_visualization()on the crop samples, specifying the CLIP model ("clip-vit-base32-torch") and abrain_keyto store the results. This function computes embeddings for each crop and projects them into a low-dimensional space (e.g., 2D) for visualization and interactive exploration in the FiftyOne App.
[ ]:
import fiftyone.brain as fob
# Get all the crop samples from the dataset
flattened_crops = grouped_dataset.select_group_slices("crop")
# Compute embeddings and visualization
results = fob.compute_visualization(
flattened_crops,
brain_key="crop_embedding_viz",
model="clip-vit-base32-torch"
)
[ ]:
session2 = fo.launch_app(flattened_crops, port=5152, auto=False)
Summary#
You explored your data and model predictions in FiftyOne
You evaluated performance and inspected edge cases
You identified concrete next steps to improve data/model quality