Pgvector Vector Search Integration#
Pgvector is a vector search extension to PostgreSQL, one of the most popular open source databases, and we’ve made it easy to use Pgvector on your computer vision data directly from FiftyOne!
Follow these simple instructions to get started using Pgvector + FiftyOne.
FiftyOne provides an API to create Pgvector indexes, upload vectors, and run similarity queries, both programmatically in Python and via point-and-click in the App.
Note
Did you know? You can search by natural language using Pgvector similarity indexes!
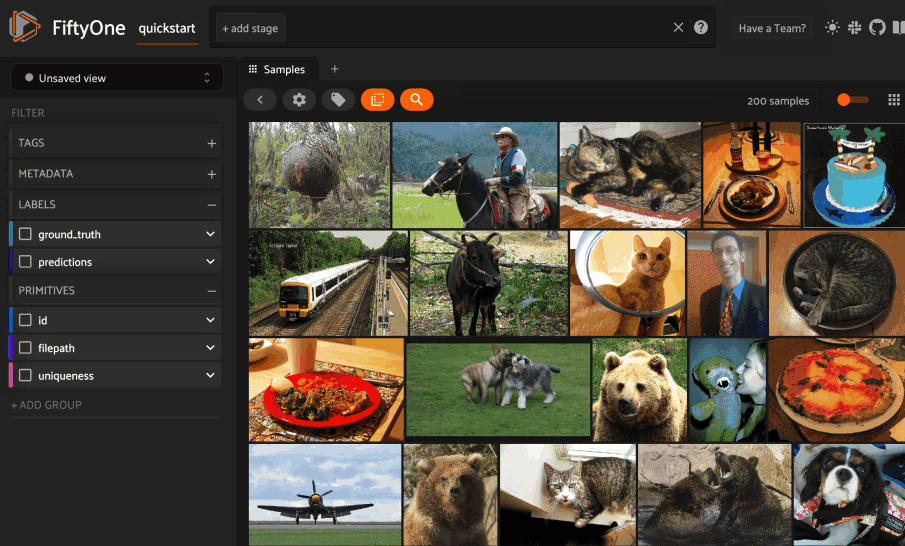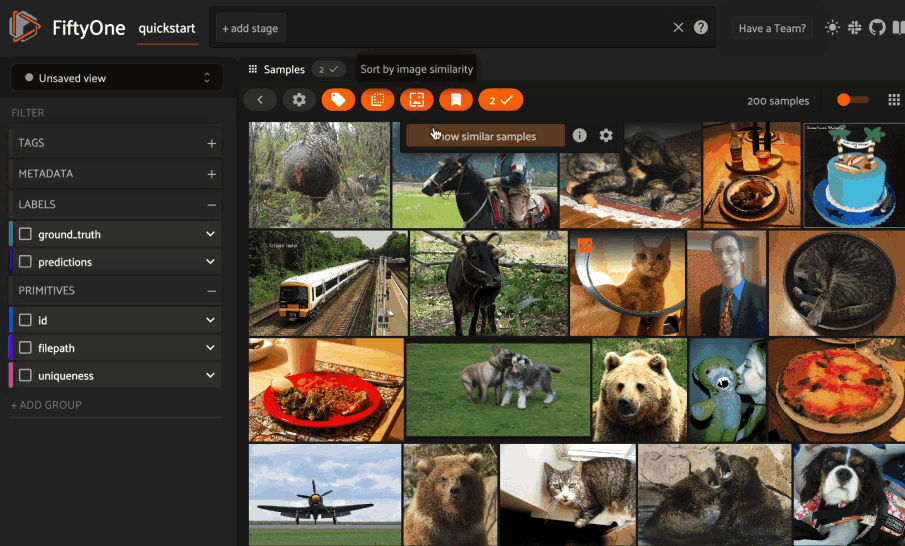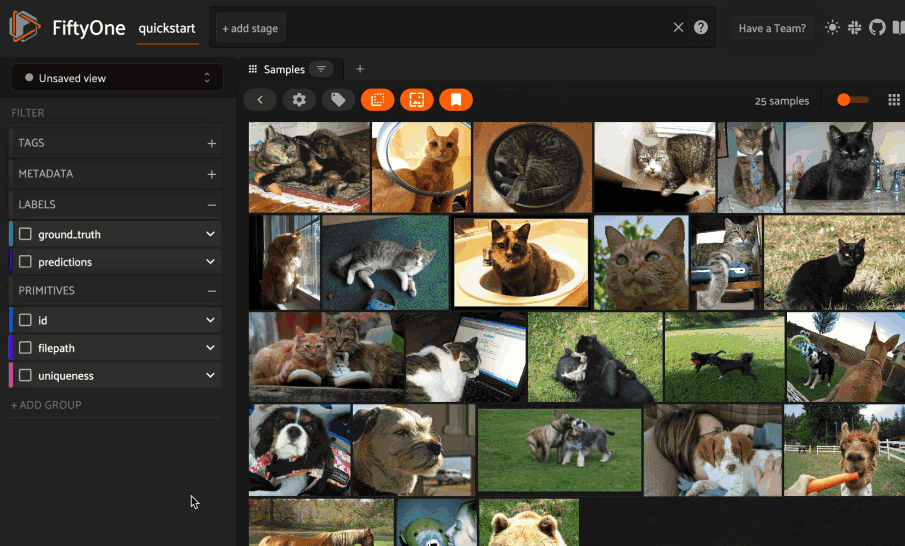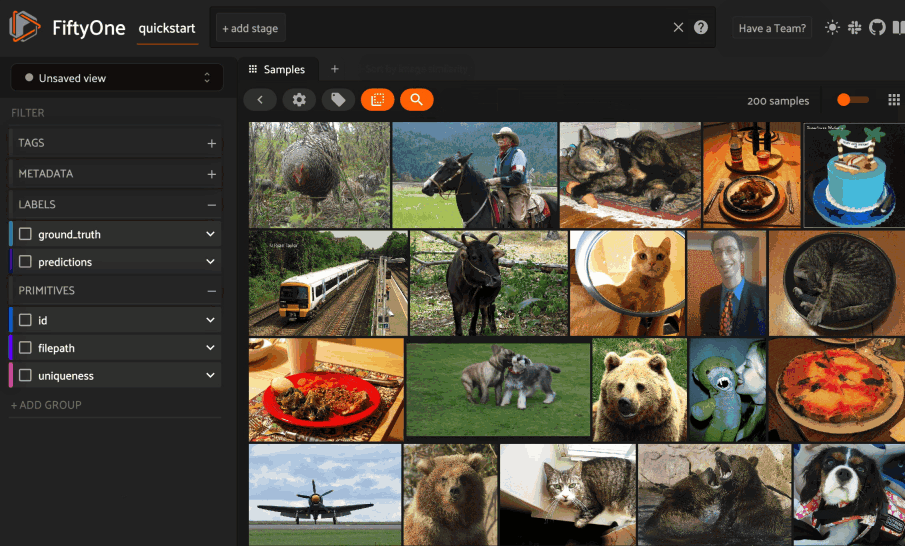
Basic recipe#
The basic workflow to use Pgvector to create a similarity index on your FiftyOne datasets and use this to query your data is as follows:
Connect to or start a PostgreSQL server with the pgvector extension
Load a dataset into FiftyOne
Compute embedding vectors for samples or patches in your dataset, or select a model to use to generate embeddings
Use the
compute_similarity()method to generate a Pgvector similarity index for the samples or object patches in a dataset by setting the parameterbackend="pgvector"and specifying abrain_keyof your choiceUse this Pgvector similarity index to query your data with
sort_by_similarity()If desired, delete the index
The example below demonstrates this workflow.
Note
You must have access to a PostgreSQL instance with pgvector extension and install the psycopg2 Python module to run this example:
pip install psycopg2
You can store credentials for your Postgres instance as described in this section to avoid entering them manually each time you interact with your Pgvector index.
First let’s load a dataset into FiftyOne and compute embeddings for the samples:
1import fiftyone as fo
2import fiftyone.brain as fob
3import fiftyone.zoo as foz
4
5# Step 1: Load your data into FiftyOne
6dataset = foz.load_zoo_dataset("quickstart")
7
8# Steps 2 and 3: Compute embeddings and create a similarity index
9pgvector_index = fob.compute_similarity(
10 dataset,
11 brain_key="pgvector_index",
12 backend="pgvector",
13)
Once the similarity index has been generated, we can query our data in FiftyOne
by specifying the brain_key:
1# Step 4: Query your data
2query = dataset.first().id # query by sample ID
3view = dataset.sort_by_similarity(
4 query,
5 brain_key="pgvector_index",
6 k=10, # limit to 10 most similar samples
7)
8
9# Step 5 (optional): Cleanup
10
11# Delete the pgvector index
12pgvector_index.cleanup()
13
14# Delete run record from FiftyOne
15dataset.delete_brain_run("pgvector_index")
Note
Skip to this section for a variety of common pgvector query patterns.
Setup#
The easiest way to get started with pgvector is to install locally via Docker.
Installing the psycopg2 client#
In order to use the pgvector backend, you must also install the psycopg2 Python module:
pip install psycopg2
Using the pgvector backend#
By default, calling
compute_similarity() or
sort_by_similarity()
will use an sklearn backend.
To use the pgvector backend, simply set the optional backend parameter of
compute_similarity() to
"pgvector":
1import fiftyone.brain as fob
2
3fob.compute_similarity(..., backend="pgvector", ...)
Alternatively, you can permanently configure FiftyOne to use the pgvector backend by setting the following environment variable:
export FIFTYONE_BRAIN_DEFAULT_SIMILARITY_BACKEND=pgvector
or by setting the default_similarity_backend parameter of your
brain config located at ~/.fiftyone/brain_config.json:
{
"default_similarity_backend": "pgvector"
}
Authentication#
If you are using a custom pgvector server, you can provide your credentials in a variety of ways.
Environment variables (recommended)
The recommended way to configure your pgvector credentials is to store them in the environment variables shown below, which are automatically accessed by FiftyOne whenever a connection to pgvector is made.
export FIFTYONE_BRAIN_SIMILARITY_PGVECTOR_CONNECTION_STRING=postgresql://postgres:mysecretpassword@localhost:5432/postgres
This is only one example of variables that can be used to authenticate a pgvector client. Find more information here.
FiftyOne Brain config
You can also store your credentials in your brain config
located at ~/.fiftyone/brain_config.json:
{
"similarity_backends": {
"pgvector": {
"connection_string": "postgresql://postgres:mysecretpassword@localhost:5432/postgres"
}
}
}
Note that this file will not exist until you create it.
Keyword arguments
You can manually provide credentials as keyword arguments each time you call
methods like compute_similarity()
that require connections to pgvector:
1import fiftyone.brain as fob
2
3pgvector_index = fob.compute_similarity(
4 ...
5 backend="pgvector",
6 brain_key="pgvector_index",
7 connection_string="postgresql://postgres:mysecretpassword@localhost:5432/postgres",
8)
Note that, when using this strategy, you must manually provide the credentials
when loading an index later via
load_brain_results():
1pgvector_index = dataset.load_brain_results(
2 "pgvector_index",
3 connection_string="postgresql://postgres:mysecretpassword@localhost:5432/postgres",
4)
pgvector config parameters#
The pgvector backend supports a variety of query parameters that can be used to customize your similarity queries. These parameters include:
index_name (None): the name of the pgvector vector search index to use or create. If not specified, a new unique name is generated automatically
table_name (None): the name of the postgres table to use or create for storing vectors. If not specified, a new unique name is generated automatically
metric (“cosine”): the distance/similarity metric to use when creating a new index. The supported values are
("cosine", "dotproduct", "euclidean", "l1", "jaccard", "hamming")work_mem (“64MB”): the base maximum amount of memory to be used by a query operation (such as a sort or hash table) before writing to temporary disk files
hnsw_m (16): The max number of connections per layer in the HNSW index
hnsw_ef_construction (64): the size of the dynamic candidate list for constructing the graph for the HNSW index
For detailed information on these parameters, see the pgvector index options documentation.
You can specify these parameters via any of the strategies described in the previous section. Here’s an example of a brain config that includes all of the available parameters:
{
"similarity_backends": {
"pgvector": {
"index_name": "your-index",
"table_name": "your-table",
"metric": "cosine",
"work_mem": "64MB",
"hnsw_m": 16,
"hnsw_ef_construction": 64
}
}
}
However, typically these parameters are directly passed to
compute_similarity() to configure
a specific new index:
1pgvector_index = fob.compute_similarity(
2 ...
3 backend="pgvector",
4 brain_key="pgvector_index",
5 index_name="your-index",
6 table_name="your-table",
7 metric="cosine",
8 work_mem="64MB",
9 hnsw_m=16,
10 hnsw_ef_construction=64,
11)
Managing brain runs#
FiftyOne provides a variety of methods that you can use to manage brain runs.
For example, you can call
list_brain_runs()
to see the available brain keys on a dataset:
1import fiftyone.brain as fob
2
3# List all brain runs
4dataset.list_brain_runs()
5
6# Only list similarity runs
7dataset.list_brain_runs(type=fob.Similarity)
8
9# Only list specific similarity runs
10dataset.list_brain_runs(
11 type=fob.Similarity,
12 patches_field="ground_truth",
13 supports_prompts=True,
14)
Or, you can use
get_brain_info()
to retrieve information about the configuration of a brain run:
1info = dataset.get_brain_info(brain_key)
2print(info)
Use load_brain_results()
to load the SimilarityIndex instance for a brain run.
You can use
rename_brain_run()
to rename the brain key associated with an existing similarity results run:
1dataset.rename_brain_run(brain_key, new_brain_key)
Finally, you can use
delete_brain_run()
to delete the record of a similarity index computation from your FiftyOne
dataset:
1dataset.delete_brain_run(brain_key)
Note
Calling
delete_brain_run()
only deletes the record of the brain run from your FiftyOne dataset; it
will not delete any associated pgvector index, which you can do as
follows:
# Delete the pgvector index
pgvector_index = dataset.load_brain_results(brain_key)
pgvector_index.cleanup()
Examples#
This section demonstrates how to perform some common vector search workflows on a FiftyOne dataset using the pgvector backend.
Note
All of the examples below assume you have configured your pgvector server as described in this section.
Create a similarity index#
In order to create a new pgvector similarity index, you need to specify
either the embeddings or model argument to
compute_similarity(). Here’s a few
possibilities:
1import fiftyone as fo
2import fiftyone.brain as fob
3import fiftyone.zoo as foz
4
5dataset = foz.load_zoo_dataset("quickstart")
6model_name = "clip-vit-base32-torch"
7model = foz.load_zoo_model(model_name)
8brain_key = "pgvector_index"
9
10# Option 1: Compute embeddings on the fly from model name
11fob.compute_similarity(
12 dataset,
13 model=model_name,
14 backend="pgvector",
15 brain_key=brain_key,
16)
17
18# Option 2: Compute embeddings on the fly from model instance
19fob.compute_similarity(
20 dataset,
21 model=model,
22 backend="pgvector",
23 brain_key=brain_key,
24)
25
26# Option 3: Pass precomputed embeddings as a numpy array
27embeddings = dataset.compute_embeddings(model)
28fob.compute_similarity(
29 dataset,
30 embeddings=embeddings,
31 backend="pgvector",
32 brain_key=brain_key,
33)
34
35# Option 4: Pass precomputed embeddings by field name
36dataset.compute_embeddings(model, embeddings_field="embeddings")
37fob.compute_similarity(
38 dataset,
39 embeddings="embeddings",
40 backend="pgvector",
41 brain_key=brain_key,
42)
Create a patch similarity index#
You can also create a similarity index for
object patches within your dataset by
including the patches_field argument to
compute_similarity():
1import fiftyone as fo
2import fiftyone.brain as fob
3import fiftyone.zoo as foz
4
5dataset = foz.load_zoo_dataset("quickstart")
6
7fob.compute_similarity(
8 dataset,
9 patches_field="ground_truth",
10 model="clip-vit-base32-torch",
11 backend="pgvector",
12 brain_key="pgvector_patches",
13)
Connect to an existing index#
If you have already created a pgvector index storing the embedding vectors
for the samples or patches in your dataset, you can connect to it by passing
the index_name to
compute_similarity():
1import fiftyone as fo
2import fiftyone.brain as fob
3import fiftyone.zoo as foz
4
5dataset = foz.load_zoo_dataset("quickstart")
6
7fob.compute_similarity(
8 dataset,
9 model="clip-vit-base32-torch", # zoo model used (if applicable)
10 embeddings=False, # don't compute embeddings
11 index_name="your-index", # the existing pgvector index
12 brain_key="pgvector_index",
13 backend="pgvector",
14)
Add/remove embeddings from an index#
You can use
add_to_index()
and
remove_from_index()
to add and remove embeddings from an existing pgvector index.
These methods can come in handy if you modify your FiftyOne dataset and need to update the pgvector index to reflect these changes:
1import numpy as np
2
3import fiftyone as fo
4import fiftyone.brain as fob
5import fiftyone.zoo as foz
6
7dataset = foz.load_zoo_dataset("quickstart")
8
9pgvector_index = fob.compute_similarity(
10 dataset,
11 model="clip-vit-base32-torch",
12 brain_key="pgvector_index",
13 backend="pgvector",
14)
15print(pgvector_index.total_index_size) # 200
16
17view = dataset.take(10)
18ids = view.values("id")
19
20# Delete 10 samples from a dataset
21dataset.delete_samples(view)
22
23# Delete the corresponding vectors from the index
24pgvector_index.remove_from_index(sample_ids=ids)
25
26# Add 20 samples to a dataset
27samples = [fo.Sample(filepath="tmp%d.jpg" % i) for i in range(20)]
28sample_ids = dataset.add_samples(samples)
29
30# Add corresponding embeddings to the index
31embeddings = np.random.rand(20, 512)
32pgvector_index.add_to_index(embeddings, sample_ids)
33
34print(pgvector_index.total_index_size) # 210
Retrieve embeddings from an index#
You can use
get_embeddings()
to retrieve embeddings from a pgvector index by ID:
1import fiftyone as fo
2import fiftyone.brain as fob
3import fiftyone.zoo as foz
4
5dataset = foz.load_zoo_dataset("quickstart")
6
7pgvector_index = fob.compute_similarity(
8 dataset,
9 model="clip-vit-base32-torch",
10 brain_key="pgvector_index",
11 backend="pgvector",
12)
13
14# Retrieve embeddings for the entire dataset
15ids = dataset.values("id")
16embeddings, sample_ids, _ = pgvector_index.get_embeddings(sample_ids=ids)
17print(embeddings.shape) # (200, 512)
18print(sample_ids.shape) # (200,)
19
20# Retrieve embeddings for a view
21ids = dataset.take(10).values("id")
22embeddings, sample_ids, _ = pgvector_index.get_embeddings(sample_ids=ids)
23print(embeddings.shape) # (10, 512)
24print(sample_ids.shape) # (10,)
Querying a pgvector index#
You can query a pgvector index by appending a
sort_by_similarity()
stage to any dataset or view. The query can be any of the following:
An ID (sample or patch)
A query vector of same dimension as the index
A list of IDs (samples or patches)
A text prompt (if supported by the model)
1import numpy as np
2
3import fiftyone as fo
4import fiftyone.brain as fob
5import fiftyone.zoo as foz
6
7dataset = foz.load_zoo_dataset("quickstart")
8
9fob.compute_similarity(
10 dataset,
11 model="clip-vit-base32-torch",
12 brain_key="pgvector_index",
13 backend="pgvector",
14)
15
16# Query by vector
17query = np.random.rand(512) # matches the dimension of CLIP embeddings
18view = dataset.sort_by_similarity(query, k=10, brain_key="pgvector_index")
19
20# Query by sample ID
21query = dataset.first().id
22view = dataset.sort_by_similarity(query, k=10, brain_key="pgvector_index")
23
24# Query by a list of IDs
25query = [dataset.first().id, dataset.last().id]
26view = dataset.sort_by_similarity(query, k=10, brain_key="pgvector_index")
27
28# Query by text prompt
29query = "a photo of a dog"
30view = dataset.sort_by_similarity(query, k=10, brain_key="pgvector_index")
Note
Performing a similarity search on a DatasetView will only return
results from the view; if the view contains samples that were not included
in the index, they will never be included in the result.
This means that you can index an entire Dataset once and then perform
searches on subsets of the dataset by
constructing views that contain the images of
interest.
Advanced usage#
As previously mentioned, you can
customize your pgvector indexes by providing optional parameters to
compute_similarity().
Here’s an example of creating a similarity index backed by a customized pgvector index. Just for fun, we’ll specify a custom index name, use dot product similarity, and populate the index for only a subset of our dataset:
1import fiftyone as fo
2import fiftyone.brain as fob
3import fiftyone.zoo as foz
4
5dataset = foz.load_zoo_dataset("quickstart")
6
7# Create a custom pgvector index
8pgvector_index = fob.compute_similarity(
9 dataset,
10 model="clip-vit-base32-torch",
11 embeddings=False, # we'll add embeddings below
12 metric="dotproduct",
13 brain_key="pgvector_index",
14 backend="pgvector",
15 index_name="custom-quickstart-index",
16)
17
18# Add embeddings for a subset of the dataset
19view = dataset.take(10)
20embeddings, sample_ids, _ = pgvector_index.compute_embeddings(view)
21pgvector_index.add_to_index(embeddings, sample_ids)