 Run in Google Colab Run in Google Colab
|
 View source on GitHub View source on GitHub
|
|
Custom Embeddings for Anomaly Detection#
In this notebook, we will explore how to generate custom embeddings for anomaly detection using the Padim model from Anomalib. Unlike general-purpose embeddings from models like CLIP or ResNet, anomaly detection requires task-specific embeddings that can distinguish between normal and abnormal samples.
Learning Objectives:#
Understand the difference between standard embeddings and anomaly-specific embeddings.
Explore how to compute embeddings using Padim from Anomalib.
Integrate these embeddings into a FiftyOne dataset.
Leverage FiftyOne for visualization and analysis.
Why Use Custom Embeddings for Anomaly Detection?#
Pre-trained models like CLIP or ResNet generate general-purpose embeddings that focus on visual similarity. However, detecting abnormalities requires learning subtle deviations from normal patterns, which these models cannot capture effectively.
Instead, we use a dedicated anomaly detection model like Padim from Anomalib, which:
Learns representations specific to normal and anomalous samples.
Extracts feature maps from an encoder (e.g., ResNet).
Compares new samples against normal feature distributions.
Further Reading:#
[ ]:
!pip install -q gdown fiftyone anomalib torch torchvision
Load the MVTec Dataset as usual#
[ ]:
import gdown
url = "https://drive.google.com/uc?id=1nAuFIyl2kM-TQXduSJ9Fe_ZEIVog4tth"
gdown.download(url, output="mvtec_ad.zip", quiet=False)
!unzip mvtec_ad.zip
import fiftyone as fo
# Define the new dataset name
dataset_name = "MVTec_AD_cEmb"
# Check if the dataset exists
if dataset_name in fo.list_datasets():
print(f"Dataset '{dataset_name}' exists. Loading...")
dataset = fo.load_dataset(dataset_name)
else:
print(f"Dataset '{dataset_name}' does not exist. Creating a new one...")
dataset_ = fo.Dataset.from_dir(
dataset_dir="/content/mvtec-ad",
dataset_type=fo.types.FiftyOneDataset
)
dataset = dataset_.clone(dataset_name)
Extracting Custom Embeddings from Padim (Anomalib)#
Instead of using a general embedding model, we will:
Load a Padim anomaly detection model using Anomalib.
Run inference on a dataset to extract anomaly embeddings.
Store the embeddings in FiftyOne for further visualization.
Relevant Documentation:
[ ]:
import torch
from anomalib.models.image.padim.torch_model import PadimModel
# Create a PaDiM model
model = PadimModel(
backbone="resnet18", # or "wide_resnet50_2", etc.
layers=["layer1", "layer2"], # choose the layers you want
pre_trained=True,
n_features=100 # optional dimension reduction
)
model.eval() # set to eval mode
[ ]:
print(model)
[ ]:
from anomalib.models.image.padim.lightning_model import Padim
import torch
from PIL import Image
import torchvision.transforms as T
# 1) Create the Lightning-based PaDiM
padim = Padim(
backbone="resnet18",
layers=["layer1", "layer2"],
pre_trained=True
)
padim.train() # so forward(...) returns embeddings
# 2) Load image
transform = T.Compose([T.Resize(224), T.ToTensor()])
# Replace this with the path to your image
image_path = "path/to/your/image.png"
pil_image = Image.open(image_path).convert("RGB")
tensor = transform(pil_image).unsqueeze(0) # (1, C, H, W)
# 3) Pass it through the model in train mode
with torch.no_grad():
embeddings = padim.model(tensor) # shape (1, embed_dim, H', W')
print(embeddings.shape)
Integrating Anomaly Embeddings into FiftyOne#
Once we obtain embeddings from Padim, we will add them to our FiftyOne dataset. This allows us to:
Perform similarity searches based on anomaly scores.
Compare normal vs. abnormal sample distributions.
Leverage FiftyOne App to inspect anomalies.
import fiftyone as fo
dataset = fo.Dataset("object_from_mvtec_ad")
# Add embeddings to each sample
for sample in dataset:
...
# Convert to CPU NumPy for storage
embedding_1d = patch_embedding.squeeze(0).cpu().numpy() # shape (D,)
# Store as a list in a new field
sample["embedding"] = embedding_1d.tolist()
sample.save()
...
Relevant Documentation: Adding Custom Fields to FiftyOne Datasets
Selecting object from MVTec AD Dataset#
[ ]:
from fiftyone import ViewField as F # helper for defining views
## get the test split of the dataset
test_split = dataset.match(F("category.label") == 'bottle')
# Clone the dataset into a new one called "mvtec_bottle"
mvtec_bottle = test_split.clone("mvtec-bottle", persistent=True)
print(mvtec_bottle)
[ ]:
print(dataset)
print(mvtec_bottle)
Calculating Embeddings using Inference with Padim Model#
[ ]:
import numpy as np
from PIL import Image
for sample in mvtec_bottle:
# Load the image via PIL
pil_image = Image.open(sample.filepath).convert("RGB")
# Apply your transform
input_tensor = transform(pil_image).unsqueeze(0) # shape (1, C, H, W)
# Compute patch embeddings in train mode
with torch.no_grad():
patch_embedding = padim.model(input_tensor) # shape (1, D, H', W')
# Optional: flatten or pool across spatial dims
# Here we use mean pooling to get a (1, D) vector
patch_embedding = patch_embedding.mean(dim=[2, 3]) # shape (1, D)
# Convert to CPU NumPy for storage
embedding_1d = patch_embedding.squeeze(0).cpu().numpy() # shape (D,)
# Store as a list in a new field
sample["embedding"] = embedding_1d.tolist()
sample.save()
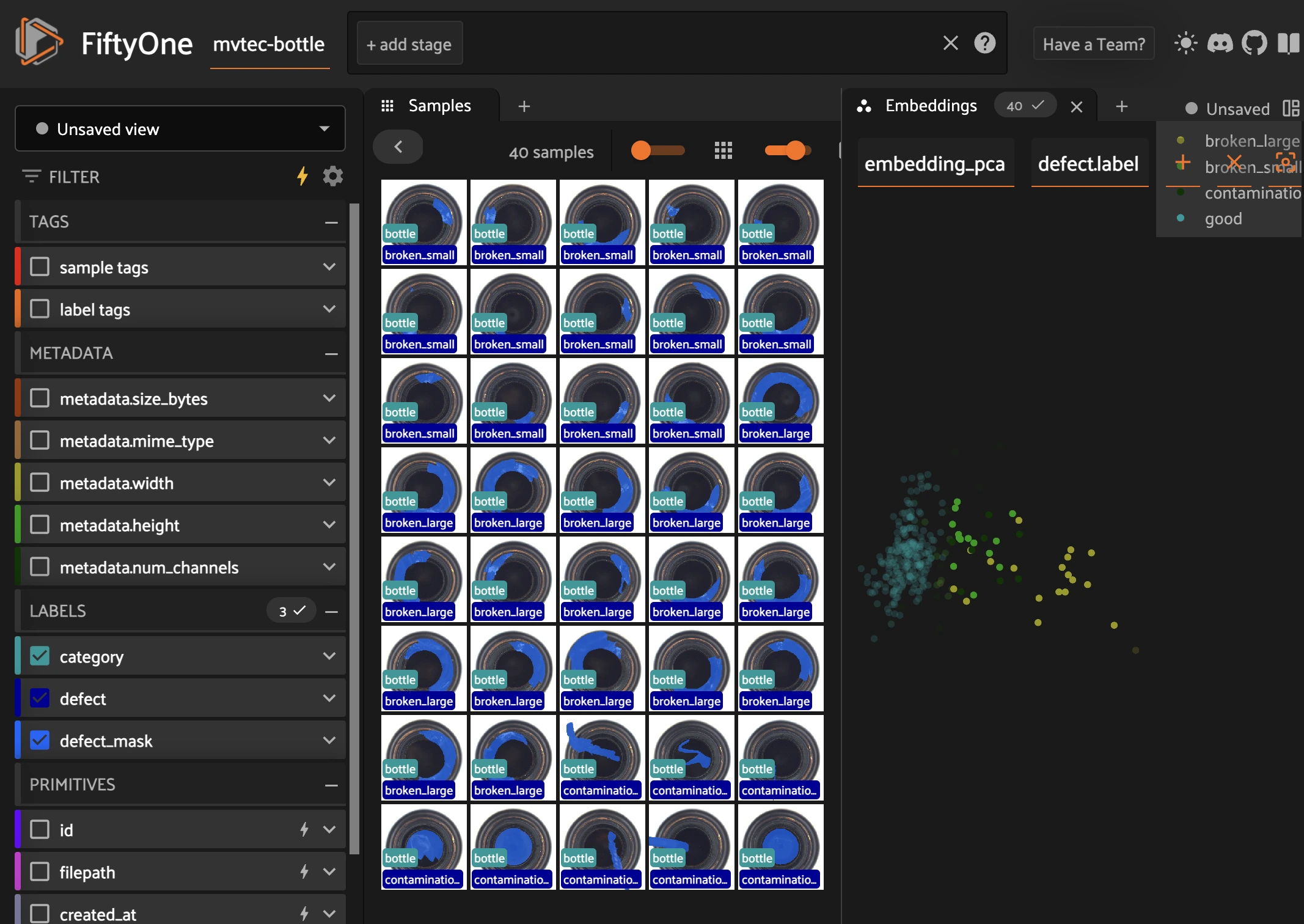
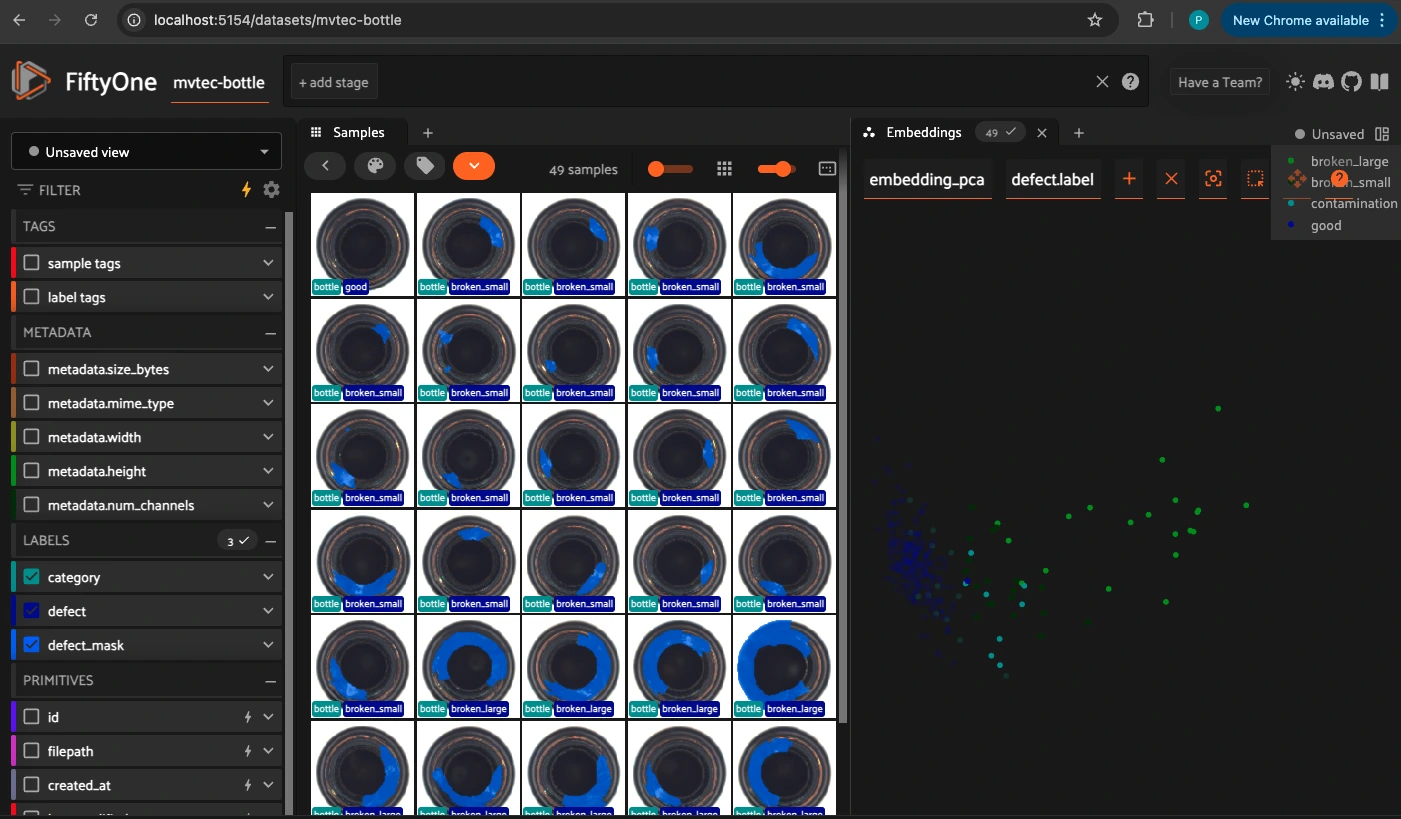
Visualizing Embeddings in FiftyOne#
[ ]:
from fiftyone.brain import compute_visualization
# This will perform PCA on the "embedding" field
compute_visualization(
mvtec_bottle,
embeddings="padin_emb",
brain_key="embedding_pca",
method="pca",
)
[ ]:
mvtec_bottle.reload()
print(mvtec_bottle)
print(mvtec_bottle.last())
[ ]:
session = fo.launch_app(mvtec_bottle, port=5154, auto=False)
print(session.url)
Next Steps:#
Try using different anomaly detection models from Anomalib and compare their embeddings with FiftyOne’s visualization tools! 🚀